Aptamer Design: From SELEX to AI-Driven Rational Design
A technical overview of modern affinity reagent design – aptamers, chemical antibodies, and computational molecular binders.
Xelari Research
•
February 2026
•
Version 1.0
00
Terminology
AptamerShort single-stranded DNA or RNA molecule that binds a specific target with high affinity. Generic term.
SELEXSystematic Evolution of Ligands by Exponential Enrichment – the traditional wet-lab method for aptamer discovery.
XelamerXelari's proprietary class of AI-designed aptamers with programmable functional features.
Affinity reagentAny molecule used to bind and detect a specific target – includes antibodies, aptamers, nanobodies, and Xelamers.
Chemical antibodyShort single-stranded DNA or RNA molecule that binds a specific target with high affinity. Generic term.
Protein binderA molecule engineered to bind a specific protein target. Aptamers and Xelamers are nucleic acid-based protein binders.
01
What are Aptamers?
[Molecular recognition by nucleic acids]
Aptamers are short single-stranded DNA or RNA oligonucleotides (typically 20–100 nucleotides) that fold into defined three-dimensional structures and bind specific molecular targets – primarily proteins – with high affinity and specificity. First described in 1990 by Tuerk & Gold and independently by Ellington & Szostak, aptamers are sometimes described as chemical antibodies – synthetic alternatives to protein-based monoclonal antibodies.
Like antibodies, aptamers recognize and bind biological targets – proteins, small molecules, cells, and even whole organisms – functioning as nucleic acid-based protein binders. Unlike antibodies, they are produced entirely by chemical synthesis, require no animals or cell cultures, exhibit superior thermal stability, and show no batch-to-batch variation. Their molecular weight (6–30 kDa) is roughly 10× smaller than a typical IgG antibody (~150 kDa), enabling better tissue penetration and more favorable pharmacokinetics.
Aptamers have demonstrated utility across diagnostics (ELISA, LFA, biosensors), therapeutics (two FDA-approved drugs: pegaptanib and avacincaptad pegol), drug delivery, proteomics, and quality control – essentially anywhere antibodies are used today.
02
The SELEX Process
[How aptamers are traditionally made]
The standard method for discovering aptamers is SELEX – Systematic Evolution of Ligands by Exponential Enrichment.
SELEX is an iterative wet-lab process:
1A random oligonucleotide library (~10¹⁵ unique sequences) is synthesized.
2The library is incubated with the target molecule.
3Non-binding sequences are washed away.
4Bound sequences are eluted and amplified by PCR.
5Steps 2–4 are repeated for 8–15 rounds over weeks to months.
The process resembles directed evolution in a test tube. Multiple SELEX variants exist – Cell-SELEX, Capture-SELEX, CE-SELEX, in vivo SELEX – each addressing specific limitations of the original protocol.
03
Why SELEX Falls Short
[Fundamental limitations after three decades of refinement]
Local Maxima Trap
A library of 10¹⁵ sequences sounds enormous, but for a 40-nt aptamer, the theoretical space is 4⁴⁰ ≈ 10²⁴. SELEX samples a vanishingly small fraction and easily gets trapped in local affinity maxima.
PCR Distortion
PCR preferentially amplifies sequences with favorable GC content, shorter length, and simpler secondary structures. Rare but potentially superior aptamers with complex folds are systematically lost.
Environment Mismatch
Aptamers selected under specific buffer, pH, temperature conditions. When transferred to real-world applications – different ionic strength, competing molecules – performance often degrades dramatically.
~30% or Below
Published reviews estimate the overall SELEX success rate at roughly 30% or below. A failed run – months of work – provides little diagnostic information about why it failed.
KEY INSIGHT
These limitations explain a paradox: aptamers as a molecular class have excellent properties, yet their commercial track record has been disappointing. The problem may not be the molecules – it may be the method used to find them.
04
The Case for Computational Aptamer Design
[Why structure prediction alone isn't enough]
AlphaFold demonstrated that AI can predict protein structures with experimental accuracy by learning from ~170,000 solved structures in PDB. Can the same approach work for aptamers?
Not directly. The Protein Data Bank contains over 200,000 protein structures, but fewer than 100 experimentally solved aptamer–protein complexes. There is simply not enough structural data to train a geometry-based neural network for aptamers the way AlphaFold was trained for proteins.
Moreover, aptamers present a fundamentally different biophysical challenge. Proteins fold into relatively rigid native states with clear thermodynamic minima. Aptamer structures are more dynamic – multiple conformations can have similar energies, and small perturbations can switch the molecule between functional and non-functional states.
DESIGN PRINCIPLE
For aptamer design, predicting geometry alone is insufficient; you need to model the energy landscape – how stable is the structure, how favorable is the binding, and how likely is the aptamer to adopt the correct conformation. Instead of learning structure from structure, learn energy from physics.
05
Xelari's Approach: Rational Aptamer Design by AI
[End-to-end computational biology platform]
Xelari has developed a computational platform that bypasses traditional SELEX workflows entirely. Instead of randomly screening billions of sequences, the platform constructs aptamers de novo using a structure-based, constraint-driven approach: the target protein's surface geometry and electrostatics define the binding problem; the platform's AI models solve it. The entire pipeline – from protein target input to ranked, ready-to-synthesize aptamer candidates – completes in 24 hours.
AliNA
Secondary Structure Prediction
Hybrid U-Net with attention mechanisms. Integrates SHAPE-seq reactivity data and coevolutionary signals. Up to 90% accuracy on non-homologous RNA, 81% on DNA (F-score, PDB test sets).
Edgar
Stability Assessment
Graph neural network estimating folding free energy (ΔG). Nucleotides as nodes, bonds as edges. R² ≈ 0.95 on RNA, R² ≈ 0.85 on DNA held-out test data.
DiNA
Target Surface Analysis & Fragment Placement
Analyzes target protein surface, selects binding sites for maximum specificity and affinity. Short oligo fragments placed using interaction physics at atomic level.
MoNA + INGA + ValentiNA
Assembly, Dynamics & Scoring
Fragments assembled (MoNA), refined via accelerated molecular dynamics (INGA). ValentiNA calculates ΔG and predicts KD for final candidate ranking.
Modifications
Chemical Modifications
2'-F, phosphorothioate backbone, biotin, Cy3/Cy5 – automated recalculation of complex stability and binding parameters.
06
Xelamers: Next-Generation AI-Designed Aptamers
[Engineered, not evolved]
The aptamers produced by the Xelari platform are called Xelamers. They belong to the same molecular class as conventional aptamers – single-stranded DNA/RNA oligonucleotides that bind specific targets – but differ in how they are created and what they can do.
Application Areas
Diagnostics & immunoassays. Drop-in replacements for antibodies in ELISA, LFA, electrochemical biosensors, and point-of-care devices. Thermal stability eliminates cold chain requirements.
Proteomics & protein detection. Scalable computational design enables rapid generation of binders against underserved targets, including the “dark proteome” – proteins for which no reliable binder currently exists.
Drug discovery tooling. Target validation binders, competitor displacement probes, and hit-generation scaffolds. A tooling layer for pharma R&D – custom affinity reagents on demand.
Quality control & environmental monitoring. Reproducible, chemically synthesized binders with defined KD values for standardized assays in manufacturing QC, food safety, and environmental testing.
07
Experimental Validation
[Independent laboratory confirmation]
The first independent validation of Xelamers was conducted by Nanohmics, Inc. (Austin, TX) against Treponema pallidum – the bacterium that causes syphilis and one of the most challenging targets in microbiology (it cannot be cultured outside a host organism).
Three Xelamers targeting three different T. pallidum surface proteins were designed in 72 hours. When synthesized and tested against live bacteria, they demonstrated dose-dependent binding and no cross-reactivity with other gram-negative organisms – confirming both affinity and specificity. Previous attempts to develop aptamers against T. pallidum via SELEX had been unsuccessful.
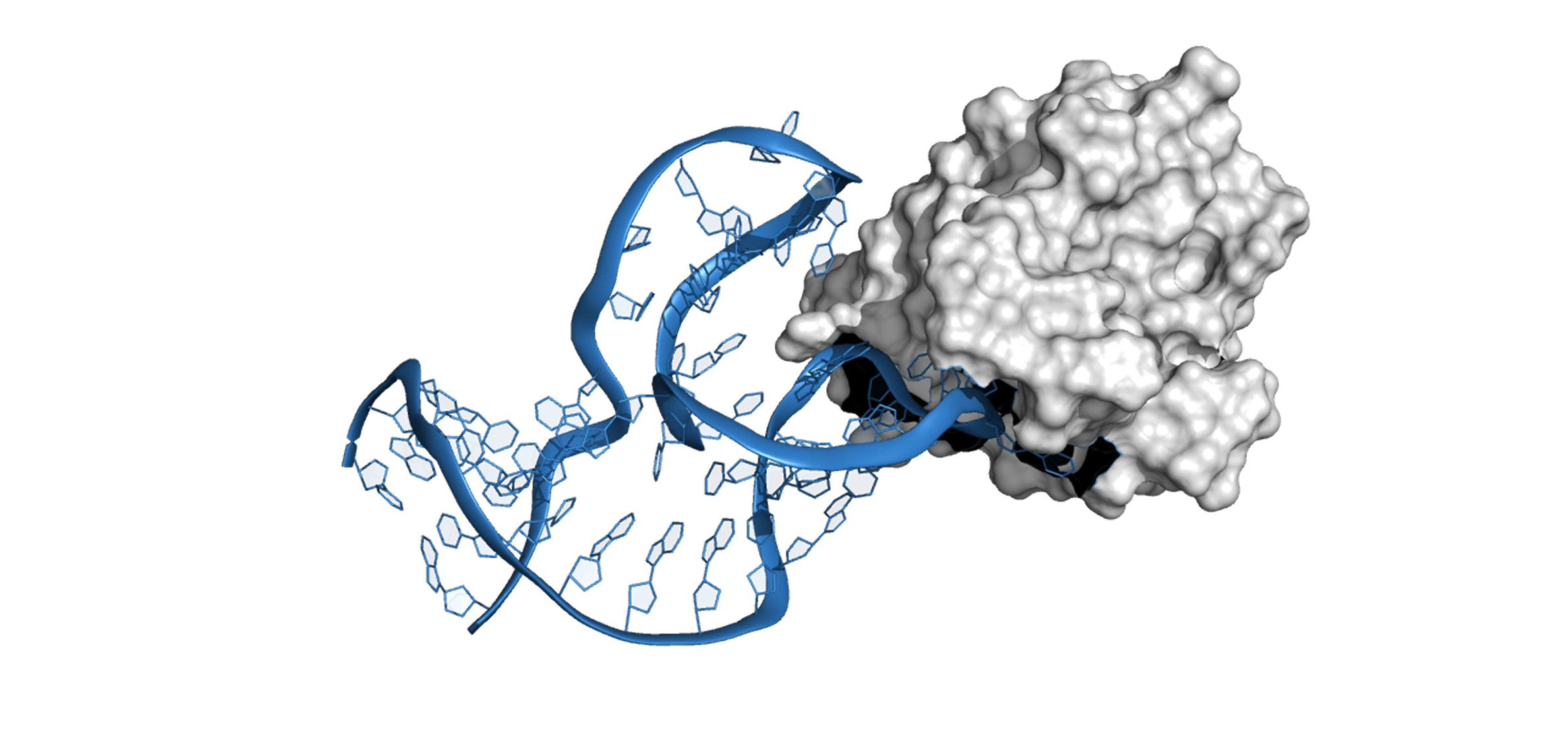
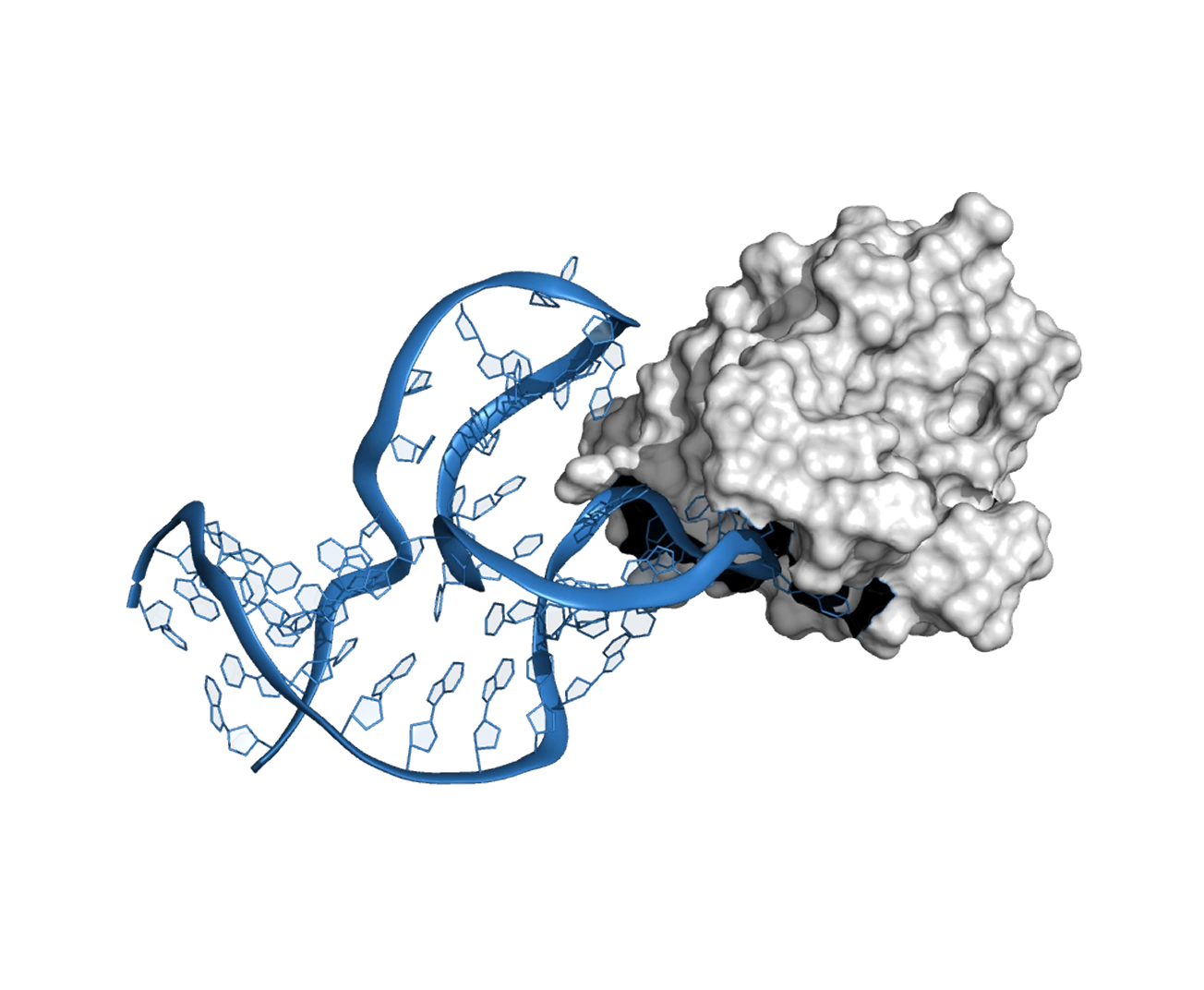
08
Frequently Asked Questions
[Common questions about aptamer design and Xelamers]
What is a chemical antibody
“Chemical antibody” is a term sometimes used for aptamers – synthetic DNA or RNA molecules that bind targets with antibody-like affinity and specificity. Unlike protein antibodies, chemical antibodies are produced by chemical synthesis, exhibit superior thermal stability, require no cold chain storage, and show no batch-to-batch variation. Xelamers are next-generation chemical antibodies: AI-designed aptamers with programmable binding properties, delivered in 24 hours.
How does Xelari's approach compare to SELEX?
SELEX requires 1–6 months of iterative wet-lab selection with variable success rates (~30% or lower). The Xelari Platform bypasses SELEX entirely through computational rational design – analyzing target protein structure and constructing aptamers with predefined affinity and specificity in 24 hours. No library screening, no PCR amplification, no sequencing.
Can AI really design an aptamer without any wet-lab work?
The design is fully computational. However, experimental validation – synthesizing the aptamer and confirming binding in the lab – remains essential. What changes is that you enter the lab with high-confidence candidates rather than random libraries, dramatically improving success rates and reducing time-to-result.
What is the difference between an aptamer and a Xelamer?
A Xelamer is an aptamer – specifically, a next-generation aptamer designed by AI on the Xelari Platform. While conventional aptamers are discovered through SELEX, Xelamers are rationally engineered using computational methods (molecular docking, energy calculation, agentic AI orchestration) and can incorporate programmable functional features beyond simple binding.
Can Xelamers replace antibodies?
In many applications, yes. For ELISA, Western blot, flow cytometry, IHC, LFA, biosensors, and point-of-care diagnostics, AI-designed aptamers offer comparable or superior binding performance with significant advantages in cost, stability, reproducibility, and development speed.
What targets can the Xelari Platform design aptamers for?
The platform accepts any protein target via UniProt accession number, PDB structure, or plain-text description. Validated against soluble proteins, membrane-associated targets, and whole-organism surface proteins (including T. pallidum). Both DNA and RNA aptamers can be designed.
Can AI-designed aptamers be used for proteomics?
Yes – one of the highest-impact applications. Current proteomics platforms are limited by affinity reagent availability. Because the platform designs aptamers in 24 hours per target, it can generate binder panels at a scale and speed impossible with SELEX or antibody development – particularly for expanding proteome coverage to include the dark proteome.
How do I design an aptamer for a membrane protein?
The platform accepts membrane protein targets in their native structural context – binding sites are selected on surface-accessible regions using PDB structures or AlphaFold-predicted models. Validated against T. pallidum outer membrane proteins where previous SELEX attempts had failed.
What inputs does the Xelari Platform need?
Three input formats: UniProt accession number (structure retrieved automatically), PDB structure file, or plain-text protein name/description. No prior aptamer data, SELEX libraries, or wet-lab results required – just the target protein.
09
References & Further Reading
[Primary sources and related publications]
- Tuerk, C. & Gold, L. (1990). Systematic evolution of ligands by exponential enrichment. Science, 249, 505–510. doi:10.1126/science.2200121
- Ellington, A.D. & Szostak, J.W. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature, 346, 818–822. doi:10.1038/346818a0
- Nasaev, S.S. et al. (2023). AliNA — a deep learning program for RNA secondary structure prediction. Molecular Informatics, 42. GitHub
- Edgar — graph neural network for nucleic acid folding energy prediction. GitHub
- Bruno, J.G., Nasaev, S., Ufaev, D. et al. Evaluation of Artificial Intelligence-Generated DNA Aptamers Against Treponema pallidum Surface Proteins. J Fluoresc (2026). doi:10.1007/s10895-026-04717-4
- Dolgusheva, V., Parfenov, N., Nasaev, S. (2025). Aptamers: Back to the Future. Biomolecula.
- Jumper, J. et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596, 583–589. doi:10.1038/s41586-021-03819-2
- Komarova, N. & Kuznetsov, A. (2019). Inside the Black Box: What Makes SELEX Better? Molecules, 24(19), 3598. doi:10.3390/molecules24193598
- Kohlberger, M. & Gadermaier, G. (2022). SELEX: Critical factors and optimization strategies. Biotechnology and Applied Biochemistry, 69(5), 1771–1792. doi:10.1002/bab.2288
Start designing now
Begin your first design today.
No SELEX. No wet lab. No guesswork.
© 2026 Xelari Inc. All rights reserved.